Last active
January 26, 2021 22:07
-
-
Save MattSkiff/9b134bf9aea4c23a673d9bc25b02ab1e to your computer and use it in GitHub Desktop.
Cluster some synthetic 1d data
This file contains hidden or bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| library(tidyverse) | |
| if (!"Ckmeans.1d.dp" %in% rownames(installed.packages())) { | |
| install.packages("Ckmeans.1d.dp") | |
| } | |
| library(Ckmeans.1d.dp) | |
| # params | |
| n1 <- 150; u1 = 1.5; sd1 = 0.2 | |
| n2 <- 1050; u2 = 4.5; sd2 = 0.6 | |
| k <- 2 # clusters | |
| cpys <- c(rnorm(n = n1, mean = u1, sd = sd1),rnorm(n = n2, mean = u2, sd = sd2)) | |
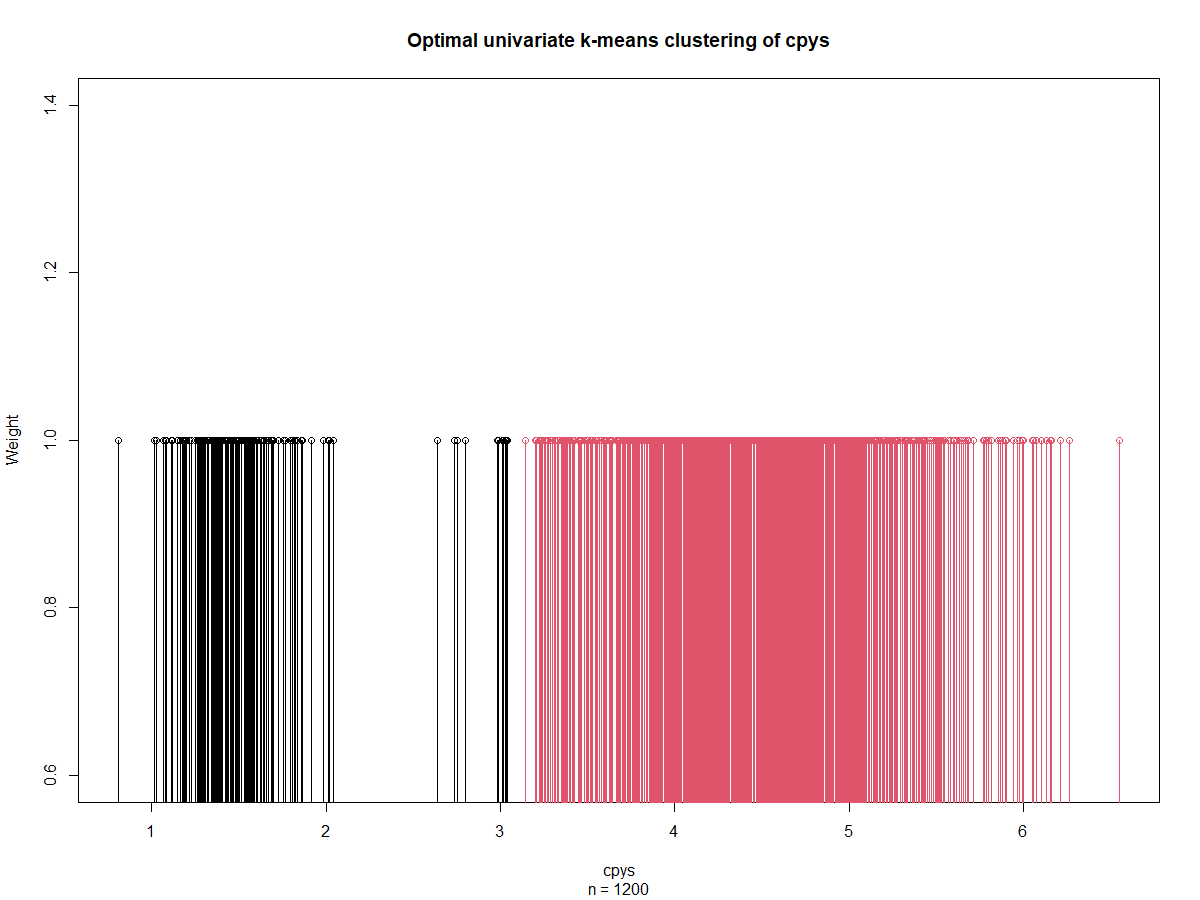
| cpy_k <- Ckmeans.1d.dp(cpys, k = k) | |
| plot(cpy_k) | |
| cpy_k.df <- data.frame(cpy = cpys,k = as.factor(cpy_k$cluster)) | |
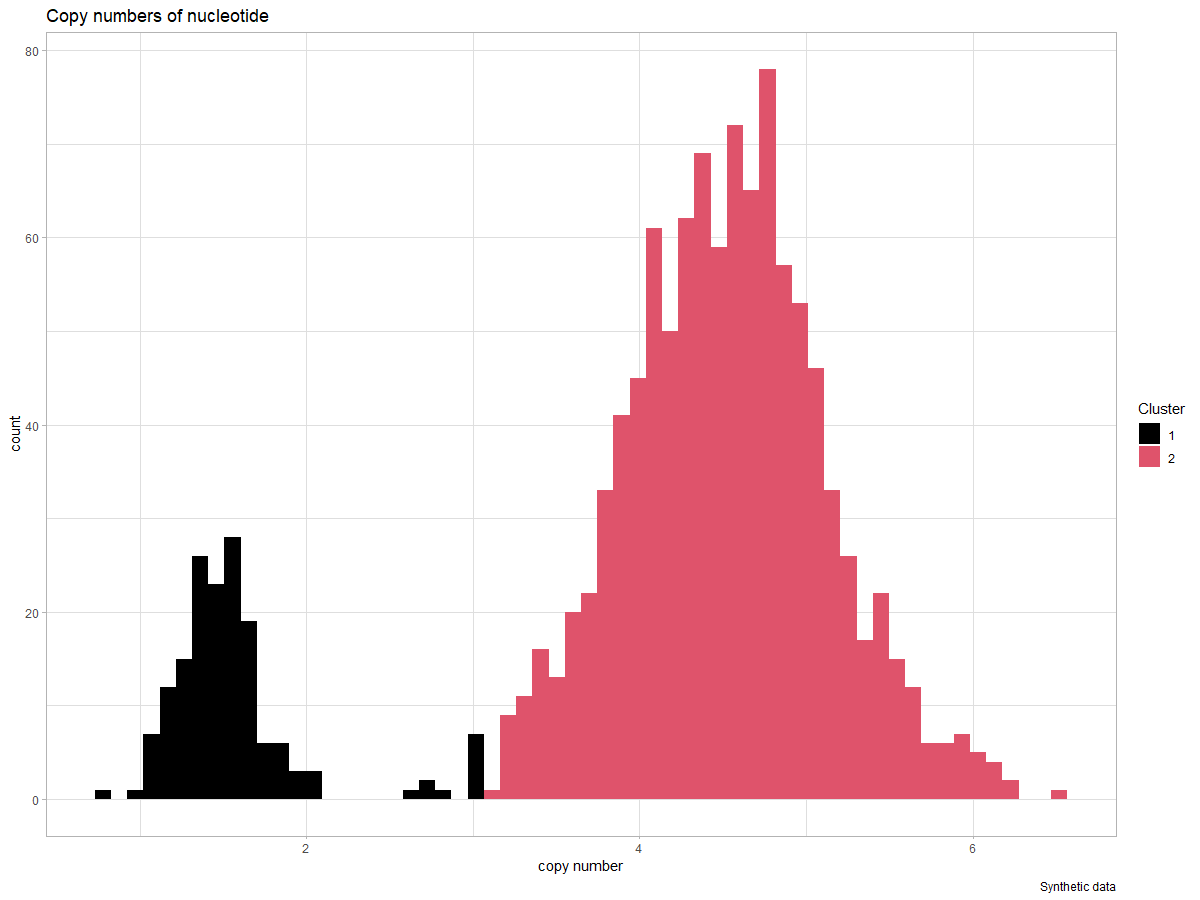
| ggplot(cpy_k.df) + | |
| geom_histogram(mapping = aes(x = cpys,group = k,fill = k),bins = 60) + | |
| theme_light() + | |
| labs(y = 'count', x = "copy number",title = "Copy numbers of nucleotide",caption = "Synthetic data") + | |
| scale_fill_manual(name = "Cluster",values = c(1,2)) | |
| # split df | |
| cpy_k_split.ls <- split(cpy_k.df, cpy_k.df$k) | |
| # range of each group | |
| lapply(FUN = function(x) { x %>% select(cpy) %>% range() },X = cpy_k_split.ls) |
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment